Surface Scan
Potential Energy Surfaces can be conveniently scanned in ASH using the calc_surface function . This function utilizes the Optimizer (from Geometry optimization) to change coordinates and carry out constrained optimizations.
This allows one to conveniently scan the potential energy surface using a convenient reaction coordinate. Both unrelaxed and relaxed scans be calculated, using either 1 and 2 reaction coordinates.
While surface scans can also be used to approximate minimum energy paths between minima and locate approximate saddlepoints ("Transition states"), it is strongly advised to instead use the Nudged Elastic Band for this purpose.
def calc_surface(fragment=None, theory=None, charge=None, mult=None, scantype='Unrelaxed', resultfile='surface_results.txt',
keepoutputfiles=True, keepmofiles=False,runmode='serial', coordsystem='dlc', maxiter=50, extraconstraints=None,
convergence_setting=None, ActiveRegion=False, actatoms=None):
Keyword |
Type |
Default value |
Details |
|---|---|---|---|
|
ASH THeory |
None |
An ASH Theory. |
|
ASH Fragment |
None |
An ASH fragment. |
|
string |
'Unrelaxed' |
What type of scan to perform. Options: 'Unrelaxed' and 'Relaxed' |
|
list of integers |
None |
List of atom indices defining Reaction coordinate(RC) 1 or 2. |
|
string |
None |
String indicating the type of reaction coordinate (either RC1 or RC2). Option: 'bond', 'angle', 'dihedral' |
|
list of floats |
None |
List of number indicating the range of values to scan for RC1 or RC2.
Example: [2.0,2.2,0.01] indicates scan value from 2.0 to 2.2 in 0.01 increments.
|
|
string |
'serial' |
Whether to run calculations in serial or parallel. Options: 'serial', 'parallel'. |
|
string |
'surface_results.txt' |
Change name of the results-file. |
|
Dict |
None |
Dictionary of additional constraints to apply during optimization. See Geometry optimization. |
|
string |
'tric' |
Which coordinate system to use during optimization.
Options: 'tric', 'hdlc', 'dlc', 'prim', 'cart'
Default: 'tric' (TRIC: translation+rotation internal coordinates),
for an active region 'hdlc' is used instead. See Geometry optimization.
|
|
integer |
100 |
Maximum number of optimization iterations before giving up (for scantype='Relaxed'). |
|
string |
None. |
Specifies the type of convergence criteria for Optimizer.
Options: 'ORCA', 'Chemshell', 'ORCA_TIGHT', 'GAU',
'GAU_TIGHT', 'GAU_VERYTIGHT', 'SuperLoose'. See Convergence section for details.
|
|
Boolean |
False |
Whether to use an Active Region during the optimization. This requires setting
the number of active atoms (actatoms list) below.
|
|
list |
None |
List of atom indices that are active during the optimization job. All other atoms are frozen. |
|
Boolean |
False |
Whether to keep outputfile for each surfacepoint from QM code or not. |
|
Boolean |
False |
Whether to keep MO-files for each surfacepoint from QM code or not. Only works for ORCATheory. |
|
Boolean |
False |
Whether to read MO-files (from mofilesdir) for each surfacepoint or not. |
|
string |
None |
Path to the directory containing MO-files. Use with read_mofiles=True option. |
|
integer |
None |
Optional specification of the charge of the system (if QM)
if the information is not present in the fragment.
|
|
integer |
None |
Optional specification of the spin multiplicity of the system (if QM)
if the information is not present in the fragment.
|
Parallelization
calc_surface is fully parallelized for both unrelaxed/relaxed and 1D/2D scans.
Use runmode='parallel' and provide as many numcores as you have available.
Make sure to turn off theory parallelization (almost never desired) by setting theory.numcores=1
How to use
The calc_surface function takes a fragment object and theory object as input. The type of scan is specified ('Unrelaxed' or 'Relaxed') and then either 1 or 2 reaction coordinates are specified via keyword arguments: RC1_type, RC1_range and RC1_indices (and RC2 versions if using two reaction coordinates).
The RC1_type/RC2_type keyword can be: 'bond', 'angle' or 'dihedral'.
The RC1_indices/RC2_indices keyword defines the atom indices for the bond/angle/dihedral. Note: ASH counts from zero.
The RC1_range/RC2_range keyword species the start coordinate, end coordinate and the stepsize (Å units for bonds, ° for angles/dihedrals).
The resultfile keyword should be used to specify the name of the file that contains the results of the scan ( format: coord1 coord2 energy). This file can be used to restart an incomplete/failed scan. If ASH finds this file in the same dir as the script, it will read the data and skip unneeded calculations. Default name : 'surface_results.txt'
calc_surface returns an ASH Results object that contains a dictionary of total energies for each surface point. The key is a tuple of coordinate value and the value is the energy, i.e. (RC1value,RC2value) : energy
1D scan:
results = calc_surface(fragment=frag, theory=ORCAcalc, scantype='Unrelaxed', resultfile='surface_results.txt',
runmode='serial', RC1_range=[180,110,-10], RC1_type='angle', RC1_indices=[1,0,2], keepoutputfiles=True)
surfacedictionary = results.surfacepoints
2D scan:
If both RC1 and RC2 keywords are provided then a 2D scan will be calculated.
results = calc_surface(fragment=frag, theory=ORCAcalc, scantype='Unrelaxed', resultfile='surface_results.txt', runmode='serial',
RC1_type='bond', RC1_range=[2.0,2.2,0.01], RC1_indices=[[0,1],[0,2]], RC2_range=[180,110,-10],
RC2_type='angle', RC2_indices=[1,0,2], keepoutputfiles=True)
surfacedictionary = results.surfacepoints
NOTE: It is possible to have each chosen reaction coordinate apply to multiple sets of atom indices by specifying a list of lists. In the 2D scan example above, the RC1_indices keyword (a 'bond' reaction coordinate) will apply to both atoms [0,1] as well as [0,2]. This makes sense when preserving symmetry of a system e.g. the O-H bonds in H2O.
Other options to calc_surface:
coordsystem (for geomeTRICOptimizer, default: 'dlc'. Other options: 'hdlc' and 'tric')
maxiter (for geomeTRICOptimizer,default : 50)
extraconstraints (for geomeTRICOptimizer, default : None. dictionary of additional constraints. Same syntax as constraints in geomeTRICOptimizer)
convergence_setting (for geomeTRICOptimizer, same syntax as in geomeTRICOptimizer)
keepoutputfiles (Boolean, keep outputfiles for each point. Default is True. )
keepmofiles (Boolean, keep MO files for each point in a directory. Default is False.)
Note: See Geometry optimization for geomeTRICOptimizer-related features.
Working with a previous scan from collection of XYZ files
If a surface scan has already been performed (by calc_surface or something else), it's possible to use the created XYZ-files and calculate single-point energies or optimizations for each surfacepoint with any level of theory.
def calc_surface_fromXYZ(xyzdir=None, theory=None, dimension=None, resultfile=None, scantype='Unrelaxed',runmode='serial',
coordsystem='dlc', maxiter=50, extraconstraints=None, convergence_setting=None, numcores=None,
RC1_type=None, RC2_type=None, RC1_indices=None, RC2_indices=None, keepoutputfiles=True, keepmofiles=False,
read_mofiles=False, mofilesdir=None):
"""Calculate 1D/2D surface from XYZ files
Args:
xyzdir (str, optional): Path to directory with XYZ files. Defaults to None.
theory (ASH theory, optional): ASH theory object. Defaults to None.
dimension (int, optional): Dimension of surface. Defaults to None.
resultfile (str, optional): Name of resultfile. Defaults to None.
scantype (str, optional): Tyep of scan: 'Unrelaxed' or 'Relaxed' Defaults to 'Unrelaxed'.
runmode (str, optional): Runmode: 'serial' or 'parallel'. Defaults to 'serial'.
coordsystem (str, optional): Coordinate system for geomeTRICOptimizer. Defaults to 'dlc'.
maxiter (int, optional): Max number of iterations for geomeTRICOptimizer. Defaults to 50.
extraconstraints (dict, optional): Dictionary of constraints for geomeTRICOptimizer. Defaults to None.
convergence_setting (str, optional): Convergence setting for geomeTRICOptimizer. Defaults to None.
numcores (float, optional): Number of cores. Defaults to None.
RC1_type (str, optional): Reaction-coordinate type (bond,angle,dihedral). Defaults to None.
RC2_type (str, optional): Reaction-coordinate type (bond,angle,dihedral). Defaults to None.
RC1_indices (list, optional): List of atom-indices involved for RC1. Defaults to None.
RC2_indices (list, optional): List of atom-indices involved for RC2. Defaults to None.
Returns:
[type]: [description]
"""
We can use the calc_surface_fromXYZ function to read in previous XYZ-files (named like this: RC1_2.0-RC2_180.0.xyz for a 2D scan and like this: RC1_2.0.xyz for a 1D scan). These files should have been created from calc_surface already (present in surface_xyzfiles results directory). By providing a theory level object we can then easily perform single-point calculations for each surface point or alternatively relax the structures employing constraints. The results is a dictionary like before.
#Directory of XYZ files. Can be full path or relative path.
surfacedir = '/users/home/ragnarbj/Fe2S2Cl4/PES/Relaxed-Scan-test1/SP-DLPNOCC/surface_xyzfiles'
#Calculate surface from collection of XYZ files. Will read old surface-results.txt file if requested (resultfile="surface-results.txt")
#Unrelaxed single-point job
results = calc_surface_fromXYZ(xyzdir=surfacedir, scantype='Unrelaxed', theory=ORCAcalc, dimension=2, resultfile='surface_results.txt' )
surfacedictionary = results.surfacepoints
#Relaxed optimization job. A geometry optimization with constraints will be done for each point
#The RC1_type and RC1_indices (and RC2_type and RC2_indices for a 2D scan) also need to be provided
results = calc_surface_fromXYZ(xyzdir=surfacedir, scantype='Relaxed', theory=ORCAcalc, dimension=2, resultfile='surface_results.txt',
coordsystem='dlc', maxiter=50, extraconstraints=None, convergence_setting=None,
RC1_type='bond', RC1_indices=[[0,1],[0,2]], RC2_type='angle', RC2_indices=[1,0,2])
surfacedictionary = results.surfacepoints
Other options:
keepoutputfiles=True (outputfile for each point is saved in a directory. Default True)
keepmofiles=False (Boolean, MO-file for each point is saved in a directory. Default False)
read_mofiles=False (Boolean: Read MO-files from directory if True. Default False.)
mofilesdir=path (Directory path containing MO-files (GBW files if ORCA) )
ActiveRegion= True/False
actatoms=list (list of active atoms if doing relaxed scan)
Parallelization
calc_surface_fromXYZ is fully parallelized.
Use runmode='parallel' and provide as many numcores as you have available.
Make sure to turn off theory parallelization (almost never desired) by setting theory.numcores=1
Plotting
The final result of the scan can be found as dictionary in the ASH Results object (returned by calc_surface and calc_surface_fromXYZ ) and can be easily plotted by giving the dictionary as input to plotting functions (based on Matplotlib). See Plotting) page.
The dictionary has the format: (coord1,coord2) : energy for a 2D scan and (coord1) : energy for a 1D scan where (coord1,coord2)/(coord1) is a tuple of floats and energy is the total energy as a float.
A dictionary using data from a previous job (stored e.g. in surface_results.txt) can be created via the read_surfacedict_from_file function:
surfacedictionary = read_surfacedict_from_file("surface_results.txt")
#For 1D scan:
#reactionprofile_plot(surfacedictionary, finalunit='kcal/mol',label='Plotname', x_axislabel='Angle', y_axislabel='Energy',
#imageformat='png', RelativeEnergy=True, pointsize=40, scatter_linewidth=2, line_linewidth=1, color='blue')
#For 2D scan:
contourplot(surfacedictionary, finalunit='kcal/mol',label="Plotname", interpolation='Cubic', x_axislabel='Bond (Å)', y_axislabel='Angle (°)')
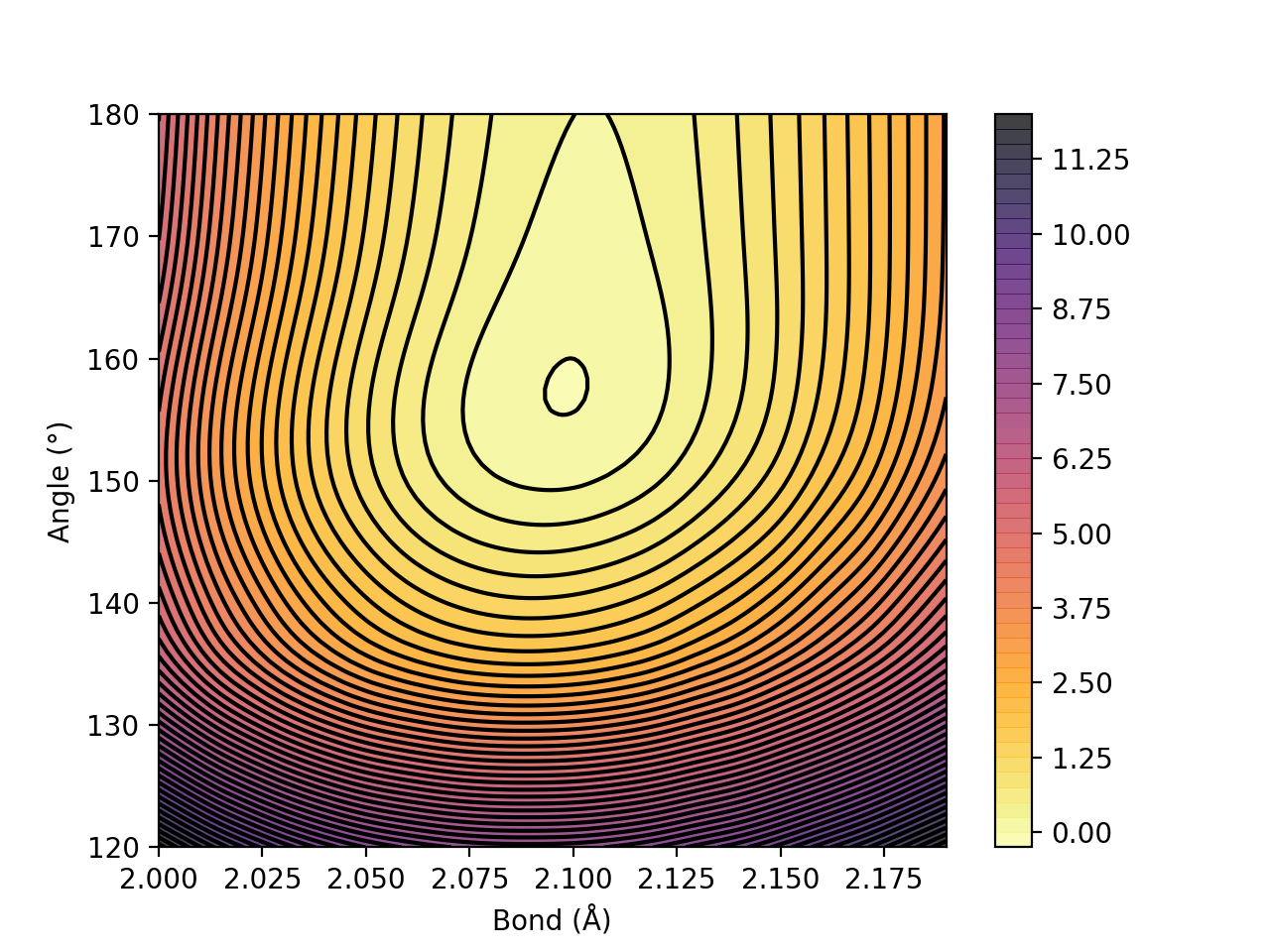
Figure. Energy surface of FeS2 scanning both the Fe-S bond and the S-Fe-S angle. The Fe-S reaction coordinate applies to both Fe-S bonds.