Electronic structure analysis
ASH contains some basic functionality for electronic structure analysis that can be useful.
NOTE: Some functions on this page requires you to import the functions_elstructure module:
from ash.functions.functions_elstructure import *
Creating and modifying Gaussian Cube files
Functions to read Gaussian Cube file,
#Read Cube file into dictionary
def read_cube(cubefile):
#Write Cube dictionary into file
def write_cube(cubedict, name="Default"):
#Subtract one Cube-file from another
def write_cube_diff(cubedict1,cubedict2, name="Default"):
#Sum of 2 Cube-files
def write_cube_sum(cubedict1,cubedict2, name="Default"):
#Product of 2 Cube-files
def write_cube_product(cubedict1,cubedict2, name="Default"):
# Read cubefile. Grabs coords. Calculates density if MO
def create_density_from_orb (cubefile, denswrite=True, LargePrint=True):
How to use the functions:
from ash import *
#Read 2 Cube-files into dictionaries
mo2=read_cube("hf.mo2a.cube")
mo3=read_cube("hf.mo3a.cube")
#Subtract, sum and multiply previously read-in Cube-file dictionaries
write_cube_diff(mo2,mo3, name="MO_2-3-diff") #Creates file MO_2-3-diff.cube
write_cube_sum(mo2,mo3, name="MO_2-3-sum") #Creates file MO_2-3-sum.cube
write_cube_product(mo2,mo3, name="MO_2-3-prod") #Creates file MO_2-3-prod.cube
# Create density from MO from a Cube-file.
create_density_from_orb("hf.mo2a.cube") #Creates file hf.mo2a-dens.cube
Create Cubefiles from orbital files
It is also possible to directly create Cubefiles from wavefunction files. If an ORCA GBW file (containing the SCF wavefunction), ORCA natural orbital file or a Molden file (e.g. created by CFOUR or MRCC) is provided then create_cubefile_from_orbfile can directly create a Cubefile from the associated WF. The function recognized the file type from the file extension and creates the Cubefile accordingly with the help of the orca_2mkl program (requires ORCA to be installed), and Multiwfn via the Multiwfn-ASH interface (See Multiwfn interface for details).
def create_cubefile_from_orbfile(orbfile, grid=3, delete_temp_molden_file=True, printlevel=2):
Create difference density from 2 Cubefiles
A more convenient option than using the write_cube_diff function above. The diffdens_of_cubefiles function reads 2 Cube-files directly and creates a difference density Cubefile.
def diffdens_of_cubefiles(ref_cubefile, cubefile):
Create multiple difference densities from all orbital-files in directory
Sometimes one would like to conveniently create difference densities from all orbital/WF files in a directory. The diffdens_tool function can be used for this purpose. It reads all files in a directory with a given extension (e.g. .gbw, .molden, .nat) and creates difference densities w.r.t. to a reference file (e.g. HF.gbw).
This function use Multiwfn via the Multiwfn-ASH interface (See Multiwfn interface for details).
def diffdens_tool(reference_orbfile="HF.gbw", dir='.', grid=3, printlevel=2):
Example: Vertical ionization of Cobaltocene
An example might be to create a difference density plot between two redox states of a molecule. This can only cleanly be done for a vertical redox process.
from ash import *
import shutil
string="""
Co 6.344947000 -1.560817000 5.954256000
C 6.026452000 -0.546182000 7.802276000
C 5.793563000 -1.965872000 7.908267000
C 7.027412000 -2.657637000 7.660083000
C 7.976024000 -1.681400000 7.254253000
C 7.360141000 -0.371346000 7.359546000
H 5.287260000 0.234934000 7.955977000
H 4.853677000 -2.430623000 8.196105000
H 7.174908000 -3.733662000 7.680248000
H 7.845323000 0.573837000 7.130600000
C 7.003675000 -1.909758000 4.002507000
C 6.025582000 -2.892708000 4.310293000
C 4.831240000 -2.191536000 4.690416000
C 5.029751000 -0.780020000 4.468693000
C 6.380790000 -0.601338000 4.083942000
H 8.038293000 -2.097559000 3.727053000
H 6.179063000 -3.967316000 4.350882000
H 3.905308000 -2.652860000 5.025216000
H 6.875603000 0.346060000 3.887076000
H 4.297348000 0.002948000 4.644348000
H 8.999375000 -1.871686000 6.940915000
"""
#Defining fragment for redox reaction
Co_neut=Fragment(coordsstring=string, charge=0, mult=2)
Co_ox=Fragment(coordsstring=string, charge=1, mult=1)
label="Cocene_"+'_'
#Defining QM theory as ORCA here
qm=ORCATheory(orcasimpleinput="! BP86 def2-SVP tightscf notrah")
#Run neutral species with ORCA
e_neut=Singlepoint(theory=qm, fragment=Co_neut)
shutil.copyfile(qm.filename+'.gbw', label+"neut.gbw") # Copy GBW file
#Run orca_plot to request electron density creation from ORCA gbw file
run_orca_plot(label+"neut.gbw", "density", gridvalue=80)
#Run oxidized species with ORCA
e_ox=Singlepoint(theory=qm, fragment=Co_ox)
shutil.copyfile(qm.filename+'.gbw', label+"ox.gbw") # Copy GBW file
#Run orca_plot to request electron density creation from ORCA gbw file
run_orca_plot(label+"ox.gbw", "density", gridvalue=80)
#Read Cubefiles from disk.
neut_cube_data = functions.functions_elstructure.read_cube(label+"neut.eldens.cube")
ox_cube_data = functions.functions_elstructure.read_cube(label+"ox.eldens.cube")
#Write out difference density as a Cubefile
functions.functions_elstructure.write_cube_diff(neut_cube_data, ox_cube_data, label+"diffence_density.cube")
The script will output the files Cocene_neut.eldens.cube and Cocene_ox.eldens.cube that are here generated by orca_plot. The file Cocene_diffence_density.cube is generated by write_cube_diff.
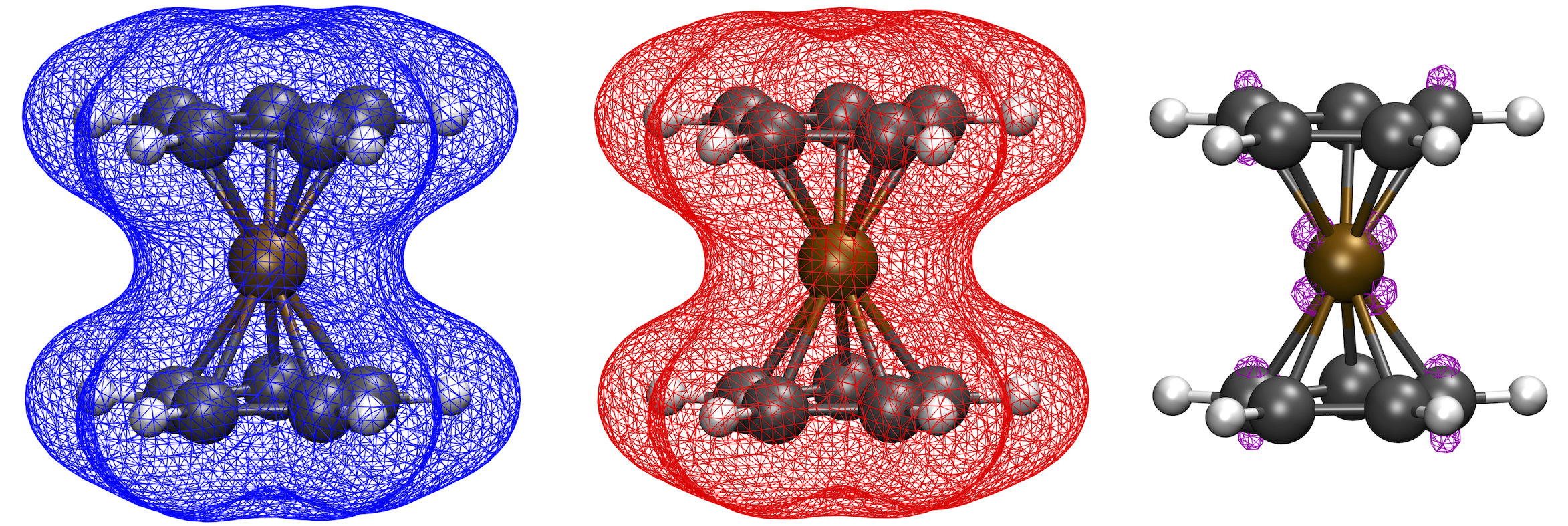
Various analysis tools
CM5 charges can be calculated using the calc_cm5 function. This function requires the atomic numbers (list), coordinates (numpy array) and Hirschfeld charges (list) of the system:
def calc_cm5(atomicNumbers, coords, hirschfeldcharges):
Functions to calculate J-couplings according to Yamaguchi, Bencini or Noodleman formulas. All functions requires the energy of the high-spin and broken-symmetry energy.
#Yamaguchi equation also requires the <S^2> values of the high-spin and BS state.
def Jcoupling_Yamaguchi(HSenergy,BSenergy,HS_S2,BS_S2):
#The Bencini equation (strong-interaction limit, i.e. bond-formation) requires the maximum spin of the system.
def Jcoupling_Bencini(HSenergy,BSenergy,smax):
#The Noodleman equation (weak-interaction limit) also requires the maximum spin of the system.
def Jcoupling_Noodleman(HSenergy,BSenergy,smax):
NOCV analysis
NOCV analysis can be performed in ASH in 2 different ways: NOCV_density_ORCA or NOCV_Multiwfn
NOCV_density_ORCA calls on ORCA to perform the NOCV and ETS-NOCV. It is unfortunately limited to closed-shell systems but the advantage is that the ETS-NOCV is performed exactly.
def NOCV_density_ORCA(fragment_AB=None, fragment_A=None, fragment_B=None, theory=None, griddensity=80,
NOCV=True, num_nocv_pairs=5, keep_all_orbital_cube_files=False,
make_cube_files=True):
The NOCV_Multiwfn function calls on Multiwfn to perform the NOCV and ETS-NOCV. The advantage is that it can be used for open-shell systems but the disadvantage is that the energy decomposition analysis is approximate as full ETS method is not performed.
def NOCV_Multiwfn(fragment_AB=None, fragment_A=None, fragment_B=None, theory=None, gridlevel=2, openshell=False,
num_nocv_pairs=5, make_cube_files=True, numcores=1, fockmatrix_approximation="ETS"):
Various ORCA-specific analysis tools
Read/write Fock matrix from/to ORCA outputfile.
# Convert Fock matrix into ORCA-format for printing. Returns string
def get_Fock_matrix_ORCA_format(Fock):
# Read Fock matrix from ORCA outputfile. Returns 2 numpy arrays (alpha and beta)
def read_Fock_matrix_from_ORCA(file):
# Write Fock matrix to disk as a dummy ORCA outputfile. Can be used by Multiwfn
def write_Fock_matrix_ORCA_format(outputfile, Fock_a=None,Fock_b=None, openshell=False):
Create difference density for 2 calculations differing in either fragment or theory-level. Theory level has to be ORCATheory. Difference density is written to disk as a Cube-file.
#Create difference density for 2 calculations differing in either fragment or theory-level
def difference_density_ORCA(fragment_A=None, fragment_B=None, theory_A=None, theory_B=None,
griddensity=80, cubefilename='difference_density'):